Workshop
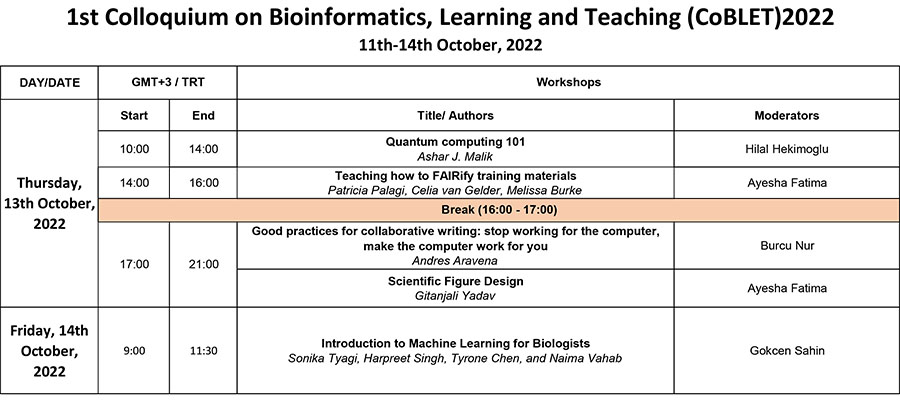
Workshops
Workshop 1
Title: Quantum computing 101
Date: 13th October, 2022
Time: 10:00-14:00 (GMT+3)
Trainer: Ashar J. Malik
About the workshop
Quantum computing is a relatively new area which promises great advancements in a variety of research fields. This workshop is designed to act as a primer, allowing the audience to interact with quantum computing environments and to introduce the basic components required to realize quantum algorithms. The workshop will make use of the Qiskit SDK. After the workshop, the audience will be able to 1) have a realistic appreciation of the current state-of-the-art of quantum computing, 2) have a basic understanding of what quantum computing is and how quantum algorithms can be realised and 3) understand cutting edge application areas in biological sciences, which can benefit from this advance, with an emphasis on quantum machine learning.
Prerequisite:The workshop will use the quantum environments made available in the IBM cloud. Before the start of the workshop the audience are required to have:
1) A modern up-to-date web browser (Safari, Mozilla Firefox, Google Chrome)
2) A suitable internet connection
3) An account on IBM Quantum (the account is free and can be made here: https://quantum-computing.ibm.com). Make sure to create the account before the start of the workshop activities.
Please register here to attend the workshops
Workshop 2
Title: Teaching how to FAIRify training materials
Date: 13th October, 2022
Time: 14:00-16:00 (GMT+3)
Trainers: Patricia Palagi, Celia van Gelder, Melissa Burke
About the workshop:
A FAIR hands-on lesson on how to FAIRify training materials was designed and initially created during events held in 2021, in particular at the ELIXIR-GOBLET workshop (“Developing (FAIR) training materials for a lesson on how to develop (FAIR) training materials”) of the GOBLET AGM 2021 and at the European BioHackathon in November 2021. Participants in these events and other colleagues from the FAIR training community (in- and outside ELIXIR and GOBLET) have continued to develop the lesson, turning it into a handbook which today counts eleven chapters.
The goals of this workshop are to
i) update the community on the current status of the handbook,
ii) share experiences on how to teach the FAIRification of training material,
iii) exchange ideas on how to improve the content, and
iv) plan the next steps towards teaching the handbook content. The workshop will be structured around the following:
A short presentation for the newcomers to introduce the topic of training material FAIRification, the current status of the handbook and its chapters.
Breakout groups to
1) share experiences,
2) discuss the best ways to teach the content - for instance the most critical parts that need to be taught in a given timeframe, teaching mode (in person, self-learning, etc.) or audience, and
3) design a lesson plan to effectively teach this content.
The outcomes of this workshop will be fed into the dedicated project of the European BioHackathon in November 2022 (Project 32: Training booster: developing FAIR training materials and Learning Paths) in which the handbook will be used to guide people through the FAIRification of their training materials.
Please register here to attend the workshops
Workshop 3
Title: Good practices for collaborative writing: stop working for the computer, make the computer work for you
Date: 13th October, 2022
Time: 17:30 – 21:00 (GMT+3)
Trainers: Andres Aravena
About the workshop:
Writing is an essential step in the Scientific Method. Without writing, there are no papers, thesis, reports, or even project proposals. The success of a bioinformatic application depends, in part, of the availability of good documentation. Often these documents are written by several authors, or at least one author and one supervisor.
Nevertheless, usually the only protocol is a constant interchange of Microsoft Word (c) documents, with over-optimistic names such as _thesis-last.docx_, _thesis-corrected.docx_, _thesis-last-last.docx_, etc. That strategy leads to chaos, confusion, and many sleepless nights fighting against the computer. The same time could be better used using the computer as an ally and focusing on the complex ideas that the machine cannot do.
In this workshop we present a series of _good practices_ learnt through experience and from the relevant literature. The key concept is _structure_, as in data structure, folder structure, and structured documents. Each good practice will be illustrated with one or more practical tools, some of them cloud-based, such as version control using git, reference manager using Zotero, structured documents using LaTeX, Overleaf, Authorea, markdown, pandoc,and Google Documents. Finally, we will also explain the key ideas of reproducibility and FAIR principles.
Please register here to attend the workshops
Workshop 4
Title: Scientific Figure Design
Date: 13th October, 2022
Time: 17:30 – 21:00 (GMT+3)
Trainers: Gitanjali Yadav
About the workshop:
This course provides a practical guide to producing figures for use in reports and publications. It is a wide ranging course that looks at how to design figures to clearly and fairly represent your data, the practical aspects of graph creation and ethics of presentation. The course will use a number of example figures adapted from common analysis tools and will have exercises for students to learn the best ways of representing continuous and/or categorical data. It requires no prior coding skills or software.
Format of the Workshop:Lectures, Exercises and Presentations
Contents
15 minutes: Introductions & Ice Break. The need for Scientific Figure Design
45 minutes: Data Visualization Theory Lecture
30 minutes: Data Representation Practical- Hand held example
30 minutes: Data Representation Practical- Exercise
15 minutes: Comfort Break
45 minutes: Design Theory Lecture
15 minutes: Ethics of Data Representation - Theory
30 minutes: Ethics of Data Representation - Practical
15 minutes: Live Q & A & Closure
Pre-Requisites & Intended audience:
This tutorial is basic/introductory training. Participants should have a basic knowledge of Data types and data structures. Suitable for students and early career researchers with interest in data ethics and data representation.
Please register here to attend the workshops
Workshop 5
Title: Introduction to Machine Learning for Biologists
Date: 14th October, 2022
Time: 9:00 to 11:30 (GMT+3)
Trainers: Sonika Tyagi, Monash University, AUSTRALIA
Harpreet Singh, Hansraj Mahila Maha Vidyalaya Punjab INDIA
Tyrone Chen, and Naima Vahab, Tyagi Lab, Monash University
About the workshop:
Genomics as a data-driven discipline has attracted a number of machine learning applications in uncovering significant patterns from the DNA, RNA, or Protein sequence data. Some of the common examples are in finding genes, protein binding sites, predicting structures, and supervised classifications of binary or multi-class labels.
In this workshop we will give a brief introduction to machine learning and its applications in genomics. The workshop content includes oral talks and interactive demo sessions. We will use DNA sequence data for machine learning based classification. Though we will be doing a quick refresher of the python functions used in the tutorial, prior exposure to Python programming will be helpful.
Contents :
- Introductory lecture ……30 min
- Tokenization interactive demo ….10 min
- Break ….5 min
- Hands-on activities …1.5 h
- Break …5 min
- Recap and Q&A ….10 min
requisites :
1) No prior knowledge of ML is required
2) Basic Python programming syntax (optional)
3) Using Google colaboratory Python notebook (optional)
Learning Objectives: :
At the end of this workshop you should be able to:
i) Run python code using Google Colab notebook
ii) Have an idea of basic Machine Learning (ML) concepts
iii) Have an hands-on experience of supervised classification of binary or multi class label data
iv) Perform basic data visualisation of input data and ML model assessment metrics
Please register here to attend the workshops
